Why COVID-19 spreads more easily than SARS
A new study suggests one reason why SARS-CoV-2, the virus that causes COVID-19, is much more transmissible.
COVID-19 has caused millions of more illnesses than its predecessor, SARS, even though both diseases are caused by similar coronaviruses. Now, a new study suggests one reason why SARS-CoV-2, the virus that causes COVID-19, is much more transmissible than SARS-CoV-1, which causes SARS.
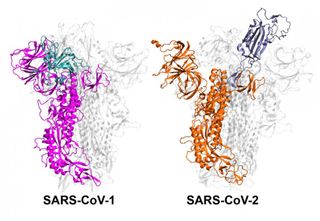
The study researchers focused on the spike protein, the structure that allows coronaviruses to bind to and enter human cells. Before either of the coronaviruses binds, it shifts its spike protein from an "inactive" to an "active" state.
Molecular simulations of these two coronaviruses suggested that SARS-CoV-2 can more easily stay in the active state and keep this position; while SARS-CoV-1 quickly alternates between the two states, which gives it less time to bind to cells.
"We discovered in these simulations that SARS-CoV-1 and SARS-CoV-2 have completely different ways of changing their shape, and on different time scales," study senior author Mahmoud Moradi, an assistant professor of physical chemistry and biochemistry at University of Arkansas, said in a statement. "SARS-CoV-1 moves faster, it activates and deactivates, which doesn't give it as much time to stick to the human cell because it's not as stable. SARS-CoV-2, on the other hand, is stable and ready to attack," said Moradi, who will present the findings, which are not yet peer-reviewed, on Thursday (Feb. 25) at the 65th Annual Meeting of the Biophysical Society, which is being held virtually this week.
Related: 20 of the worst epidemics and pandemics in history
In the year since SARS-CoV-2 emerged, it has infected more than 112 million people worldwide and is still spreading. In contrast, SARS caused a little more than 8,000 illnesses during an outbreak in 2003, but it was contained before spreading further, with the last cases reported in 2004, according to the Centers for Disease Control and Prevention.
While many studies have focused on the binding of the spike protein to human cells, relatively few have looked at the spike protein's transition between the active and inactive states.
Sign up for the Live Science daily newsletter now
Get the world’s most fascinating discoveries delivered straight to your inbox.
Based on the new study results, "we hypothesize that the greater propensity of the SARS-CoV-2 spike protein to remain in the active conformation contributes to the higher transmissibility of SARS-CoV-2 in comparison to SARS-CoV-1," the researchers wrote in their paper, which has been posted to the preprint database bioRxiv.
The findings also suggest that a region at the tip of the spike protein, known as the N-terminal domain (NTD), helps stabilize the spike protein. The N-terminal domain hasn't received much attention from researchers because it doesn't directly bind to human cells. But the NTD appears to be involved in the spike protein's transition from the inactive to active state, and so mutations in the region could affect transmissibility, the researchers said.
The results may also have implications for future therapeutics for COVID-19. "We could design therapeutics that alter the dynamics [of the spike protein] and make the inactive state more stable, thereby promoting the deactivation of SARS-CoV-2. That is a strategy that hasn't yet been adopted," Moradi said in the statement.
Originally published on Live Science.

Rachael is a Live Science contributor, and was a former channel editor and senior writer for Live Science between 2010 and 2022. She has a master's degree in journalism from New York University's Science, Health and Environmental Reporting Program. She also holds a B.S. in molecular biology and an M.S. in biology from the University of California, San Diego. Her work has appeared in Scienceline, The Washington Post and Scientific American.
